Mpacts in press
15-12-2016
A publication based on simulations performed using the Mpacts software platform recently appeared in the high ranking journal PNAS. By considering cells as 'self-propelled particles' which actively move and adhere to each other, a phase diagram was constructed showcasing various structural and dynamic organizations of epithelial cells and cell monolayers.
https://dx.doi.org/10.1073/pnas.1521151113
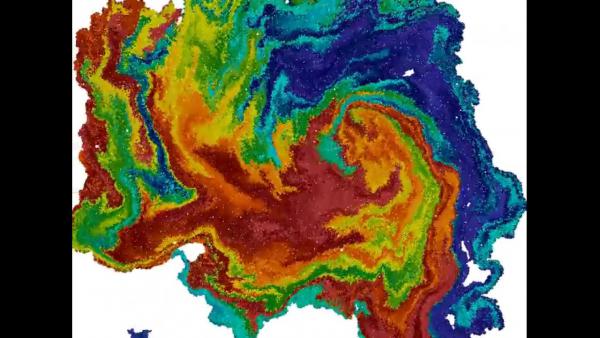
Interestingly, it was shown that the interplay between cell-cell adhesion (effected by contractile forces) and contact inhibition of locomotion can give rise to complex patterns and emergent structures. For example, grid-like phases may appear, as well as large continuous monolayers which are collectively migrating. This movie shows a simulation of a dense two-dimensional cell colony which, after structural coarsening, exhibits large-scale collective movement and interesting, fractal-like structural patterns.
New feature: Unit system
1-12-2016
Shortly, Mpacts will introduce a generic unit system for its particle-based simulations. Any physical property that is used in a simulation can be given an optional 'unit', which is passed to the underlying C++ code. Automatic unit conversions are provided. Moreover, all the used units will be checked for internal consistency. For example, in the 'gravity' command, the units of the property 'g' must be provided as being m/s^2. Otherwise, its product with the particle's mass will not result in a valid force in Newton.
Improved documentation
1-12-2016
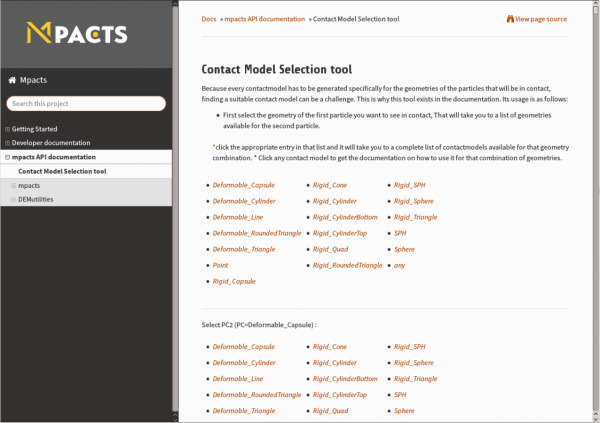
In the latest major changes of Mpacts, the documentation was given a whole new look and feel. It now has a more intuitive and easier-to-use style in line with the Mpacts design. Also under the hood, it saw some important changes. Contact models now present a convenient table that lists all possible geometry combinations for which the model can be applied. Moreover, we now also provide a "Contact Model Selection Tool". With this handy tool, you can find out which contact models make sense for the combination of geometries you want to use in your simulation.
Mpacts in press
20-10-2016
A recent paper in the journal PLOS Computational Biology saw use of the Mpacts code combined with Finite Element Method simulations in FreeFEM++.
http://dx.doi.org/10.1371/journal.pcbi.1005108
The Immersed Boundary Method was used to couple a deformable cell model described in Mpacts with an Eulerian description for the fluid domain. With this model, the mechanical influence of perfusion flow on hPDCs seeded on the surface of a titanium scaffold was investigated. The mechanical description of the cell was calibrated based on Atomic Force Microscopy measurements, combined with simulations of Micropipette Aspiration. Simulations of flow in a scaffold pore showed the distribution of shear stress, cortical tension and pressure over the surface of the cell, and investigated effects of shielding when considering small clusters of cells.