DEM explanation
This video gives a short explanation of a simple DEM simulation. A fundamental difference with the Finite Element Method is that there is no subdivision of space. 'Primitive' particles (in this case spheres) are explicitly represented by their position, radius and orientation.
In the first step of the method, all forces acting on the particles are collected. These may be external forces (called body forces) such as gravity, but the real complexity of DEM lies in the handling of contacts. Contacts are resolved based on geometric information such as overlap, or contact area and based on a "contact force model", such as Hertz' law, in which geometrical contact properties are translated into mechanical forces.In the example, this happens for both sphere-sphere contacts but also for contacts between the spheres and their surrounding container.
In a next step, the forces are summed to a single force and a single moment acting at the particle's center of gravity. Since we now know all forces acting on the particle but also know its mass and inertia, we can predict the acceleration for a given time step. By repeating this process a lot and integrating the acceleration to a new position and velocity, we obtain a dynamic simulation.
Polymer physics
The mechanical properties of living systems are often determined by complex polymer networks, for example the extracellular matrix (ECM) and the cell's cytoskeleton. Due to their highly nonlinear properties, anisotropy and active characteristics, the dynamics and bulk mechanical behavior of these networks are still not very well understood. Using the Mpacts software, we can simulate the dynamics of cytoskeletal constituents, modeled as semi-flexible polymers. This approach is computationally cheaper than classical 'bead-spring' models, yet compared to 'rigid rods' still permits us to capture the deformations caused by molecular motors on single polymers, and fluctuations on a scale comparable to the characteristic length of a full network. Molecular motors (myosin) which exert active forces on the individual actin filaments can be explicitly included.
http://dx.doi.org/10.1039/C5SM03106K
Simulations like this can help unravel the complex non-equilibrium thermodynamics, the rheological behavior and the emergence of structure and patterns that occur in living cells.
Active cellular systems
Multicellular colonies can exhibit a wide range of different phenotypes. Even in in vitro systems, collections of cells can be distributed as isolated cells, assemble into continuous monolayers, form multiple layers, or even form dense three-dimensional aggregates. The characteristics of cell motility strongly influence the types the muticellular structures that might arise.
Using Mpacts, cells can be represented as 'Self-Propelled Particles' (SPP), endowed with interactions that capture generic active cell behavior. Simulations of such models have shown that the cell-type-specific contact inhibition of locomotion (CIL), coupled with cell-cell and cell-substrate adhesion, can give rise to a rich landscape of phase behavior, showcasing gel-like structures, disperse systems and even large-scale collective motion.
https://dx.doi.org/10.1073/pnas.1521151113
This video shows a simulation of cells modeled as SPP, which are influenced by autologous chemotaxis. The cells produce a chemical species, and preferentially migrate in the positive gradient of this chemoattractant. The color scale corresponds to the magnitude of this gradient. Reaction-diffusion equations are solved on a regular grid, while the cells are represented as meshless discoidal particles.
Mechanobiology
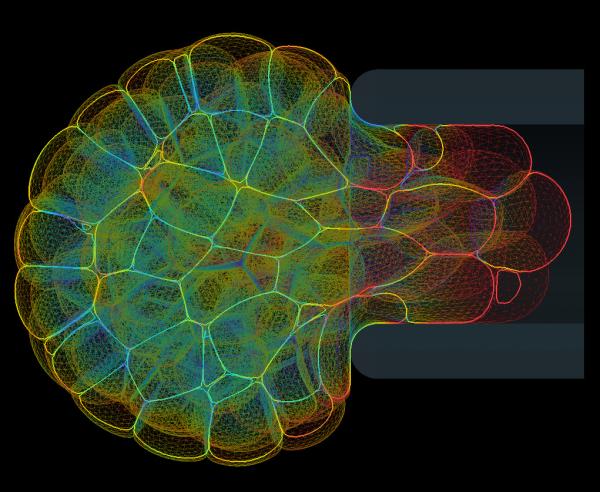
Mpacts can simulate mechanical interactions between flexible bodies composed out of triangulated surfaces. Within the DEM research group of KU Leuven, a deformable cell model was developed that captures the essence of a cell's instantaneous mechanical behavior and deformation. Moreover, intercellular adhesive and repulsive forces can be included in a mechanistic way by numerically integrating an expression for the contact pressure. This model was used to investigate the dynamics of initial cell spreading:
http://dx.doi.org/10.1371/journal.pcbi.1003267
The mechanical behavior of these deformable cells can be calibrated based on standard experimental measurements, such as Atomic Force Microscopy (AFM) and Micropipette Aspiration (MA). Moreover, the model can be used to estimate cell-cell interaction forces inside of multicellular aggregates. This image shows a simulated MA experiment, in which a a small aggregate of cells is aspirated into a pipette to measure its viscoelastic behavior. These simulations help unravel the contributions of single-cell mechanics and active and passive interaction properties to the complex rheological behavior of cell aggregates and biological tissue.
Tissue Engineering
Recent years have seen an elevated focus on tissue engineering, tissue/organ bio-printing, microfluidics and lab-on-a-chip technologies which are characterized by bottom-up development strategies and an increased importance of controlling and manipulating inter-cellular interactions.
Individual cell-based simulations in Mpacts are being used to help understand and facilitate the design of in vitro cell culture processes for tissue engineering applications. By including proper mechanical interactions between cells and their environment, simulations can be used to quantify the physical micro-environment experienced by individual cells. The properties of this micro-environment are an important determinant for later cell fate in a tissue engineering production process.
By providing a coupled DEM-CFD framework, Mpacts can be used to estimate the effects of shear flow on adherent cell cultures. This image shows a cluster of cells attached to a titanium scaffold which is placed inside a perfusion bioreactor. The immersed boundary method is used to introduce deformable cells in the fluid domain. After calibrating the mechanical properties of these cells based on Atomic Force Microscopy (AFM) and Micropipette Aspiration (MA) experiments, these simulations reveal the deformation, cortical tension and shear stress induced by the perfusion flow on the attached cells.
http://dx.doi.org/10.1371/journal.pcbi.1005108

Key publications
[1] Leblicq, T., Smeets, B., Ramon, H., Saeys, W. (2016). A discrete element approach for modelling the compression of crop stems. Computers and Electronics in Agriculture (123), 80-88. http://dx.doi.org/10.1016/j.compag.2016.02.018
[2] Smeets, B., Odenthal, T., Keresztes, J., Vanmaercke, S., Van Liedekerke, P., Tijskens, E., Saeys, W., Van Oosterwyck, H., Ramon, H. (2014). Modeling contact interactions between triangulated rounded bodies for the discrete element method. Computer Methods in Applied Mechanics and Engineering, 277, 219-238. http://dx.doi.org/10.1016/j.cma.2014.04.017
[3] Smeets, B., Odenthal, T., Vanmaercke, S., Ramon, H. (2015). Polygon-based contact description for modeling arbitrary polyhedra in the Discrete Element Method. Computer Methods in Applied Mechanics and Engineering,290, 277-289. http://dx.doi.org/10.1016/j.cma.2015.03.004
[4] Smeets, B., Odenthal, T., Tijskens, E., Ramon, H., Van Oosterwyck, H. (2013). Quantifying the Mechanical Micro-environment during Three-dimensional Cell Expansion on Microbeads by means of Individual Cell-based Modelling. Computer Methods in Biomechanics and Biomedical Engineering, 16 (10), 1071-1084. http://dx.doi.org/10.1080/10255842.2013.829461
[5] Odenthal, T., Smeets, B., Van Liedekerke, P., Tijskens, E., Van Oosterwyck, H., Ramon, H. (2013). Analysis of initial cell spreading using mechanistic contact formulations for a deformable cell model. PLoS Computational Biology, 9 (10), e1003267. http://dx.doi.org/10.1371/journal.pcbi.1003267
[6] Smeets, B., Alert, R., Pesek, J., Pagonabarraga, I., Ramon, H., Vincent, R. (2016). Emergent structures and dynamics of cell colonies by contact inhibition of locomotion. Proceedings of the National Academy of Sciences of the United States of America, 113 (51), 14621-14626. http://dx.doi.org/10.1073/pnas.1521151113
[7] Pesek, J., Baerts, P., Smeets, B., Maes, C., Ramon, H. (2016). Mathematical model suitable for efficient simulation of thin semi-flexible polymers in complex environments. Soft Matter, 12 (14), 3360-3387. http://dx.doi.org/10.1039/C5SM03106K
[8] Van Liedekerke, P., Smeets, B., Odenthal, T., Tijskens, E., Ramon, H. (2013). Solving microscopic flow problems using Stokes equations in SPH. Computer Physics Communications, 184 (7), 1686-1696. http://dx.doi.org/10.1016/j.cpc.2013.02.013
[9] Guyot, Y., Smeets, B., Odenthal, T., Subramani, R., Luyten, F., Ramon, H., Papantoniou, I., Geris, L. (2016). Immersed Boundary Models for Quantifying Flow-Induced Mechanical Stimuli on Stem Cells Seeded on 3D Scaffolds in Perfusion Bioreactors. PLoS Computational Biology, 12 (9), art.nr. e1005108. http://dx.doi.org/10.1371/journal.pcbi.1005108
[10] Diels, E., Odenthal, T., Keresztes, J., Vanmaercke, S., Verboven, P., Nicolai, B., Saeys, W., Ramon, H., Smeets, B.(2016). Development of a visco-elastoplastic contact force model and its parameter determination for apples. Postharvest Biology and Technology, 120, 157-166. http://dx.doi.org/10.1016/j.postharvbio.2016.06.003